Research
August 16, 2010 — Jeff WatsonIf you are interested in a position in the Watson lab, contact Dr. Watson by e-mail (watsonj at gonzaga) or take a look at the Gonzaga Science Undergraduate Research page and click on Gonzaga Faculty Research Interests.
In the Watson lab, we study how changes in enzyme structure affect the enzyme’s function. We use a wide range of biophysical techniques, including UV-vis, fluorescence and circular dichroism spectroscopy, as well as common molecular biology and biochemistry techniques to determine what causes an enzyme’s structure to undergo changes, and how and why those changes are important.
HMG-CoA Reductase
A major project in the lab centers on the enzyme HMG-CoA reductase (HMGR). In most organisms, this enzyme catalyzes the key regulatory step in the biosynthesis of isoprenoids, an important class of biomolecules that includes cholesterol, membrane anchor groups, plant hormones, branched fatty acids in archaeal membranes and many others. HMGR is the target of statin drugs, one of the most highly prescribed classes of drugs in the world. By inhibiting HMGR, the body is unable to make its own cholesterol and instead removes it from the bloodstream.
We, however, are particularly interested in the bacterial forms of this  enzyme. Studies have shown that, in some bacteria, HMGR is required for survival, making it an attractive potential antibiotic target. We have cloned and expressed HMGR from the opportunistic lung pathogen Burkholderia cenocepacia (right), an organism that is a leading cause of death in, for example, cystic fibrosis patients. This organism is naturally resistant to a wide range of common antibiotics, and it is therefore important to discover new antibiotic targets against it.
enzyme. Studies have shown that, in some bacteria, HMGR is required for survival, making it an attractive potential antibiotic target. We have cloned and expressed HMGR from the opportunistic lung pathogen Burkholderia cenocepacia (right), an organism that is a leading cause of death in, for example, cystic fibrosis patients. This organism is naturally resistant to a wide range of common antibiotics, and it is therefore important to discover new antibiotic targets against it.
From a biochemical perspective, bacterial HMGRs are particularly interesting because of the relatively unusual reaction it catalyzes and the necessity of a dynamic structural feature of the enzyme in making the reaction happen. Most enzymes similar to HMGR catalyze a reversible two-electron oxidation or reduction (for example, an alcohol to an aldehyde). However, HMGR catalyzes a reversible four-electron reduction (see below) without releasing an intermediate.
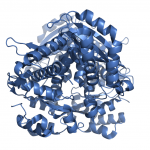 These enzymes have an important structural domain at the C-terminal end, fifty residues that are unstructured when nothing is bound to the enzyme, but form three alpha-helices that close over the active site in the presence of some substrates, but not others. The structures of the enzyme in both the open flap (above) and closed flap (below) state are shown at the left, with substrates and cofactors rendered in stick form and the flap domain colored yellow. We are particularly interested in determining how and why this domain acts as it does, and are pursuing a number of experiments to determine the mechanism by which this occurs.
These enzymes have an important structural domain at the C-terminal end, fifty residues that are unstructured when nothing is bound to the enzyme, but form three alpha-helices that close over the active site in the presence of some substrates, but not others. The structures of the enzyme in both the open flap (above) and closed flap (below) state are shown at the left, with substrates and cofactors rendered in stick form and the flap domain colored yellow. We are particularly interested in determining how and why this domain acts as it does, and are pursuing a number of experiments to determine the mechanism by which this occurs.
Stu dents in the Watson lab have a number of projects to select from, but at a minimum, all students will be involved in protein purification and enzyme kinetics experiments. Other potential projects include site-directed mutagenesis, fluorescence spectroscopy, protein folding studies, measurement of protein hydrogen-deuterium exchange rates by mass spectroscopy and x-ray crystallography studies.
dents in the Watson lab have a number of projects to select from, but at a minimum, all students will be involved in protein purification and enzyme kinetics experiments. Other potential projects include site-directed mutagenesis, fluorescence spectroscopy, protein folding studies, measurement of protein hydrogen-deuterium exchange rates by mass spectroscopy and x-ray crystallography studies.
Current projects underway:
* Kinetic characterization of B. cenocepacia HMGR: In order to compare Burkholderia cenocepacia HMGR (BcHMGR) to other previously characterized HMGRs, we must determine a number of key kinetic parameters with a variety of substrates and cofactors.
* Cysteine-scanning mutagenesis of the flap domain: A number of useful fluorescence probes modify the thiol group of cysteine residues. By mutating BcHMGR to include single cysteine residues at a number of sites on the flap domain, we can attach these probes and perform a number of fluorescence spectroscopic studies on the enzyme.
* Folding studies of BcHMGR: Circular dichroism spectroscopy is a common way to study the folding pathways of proteins. Class II HMGRs have interesting structural characteristics that make the study of their folding pathway particularly interesting, such as their oligomeric state and the inducible folding of the flap domain.
Allosteric inhibitors of bacterial carbonic anhydrase
In a collaborative project with Drs. Warren and Cronk in the GU Department of Chemistry and Biochemistry, we are studying the effects of molecules designed to be allosteric inactivators of E. coli carbonic anhydrase. The Warren lab is responsible for the design and synthesis of compounds, the Cronk lab studies the structures of the enzyme in complex with particularly interesting compounds, and the Watson lab performs kinetic analysis on the complexes. Carbonic anhydrase is known to be a key enzyme in a number of bacterial species, and those forms that have the allosteric site are significantly different than the forms expressed in humans, making the bacterial enzyme an attractive target for novel antibiotics.
If you are interested in work related to this project, please contact Dr. Watson directly.